Affinity Proteomics
Proximity Extension Assay
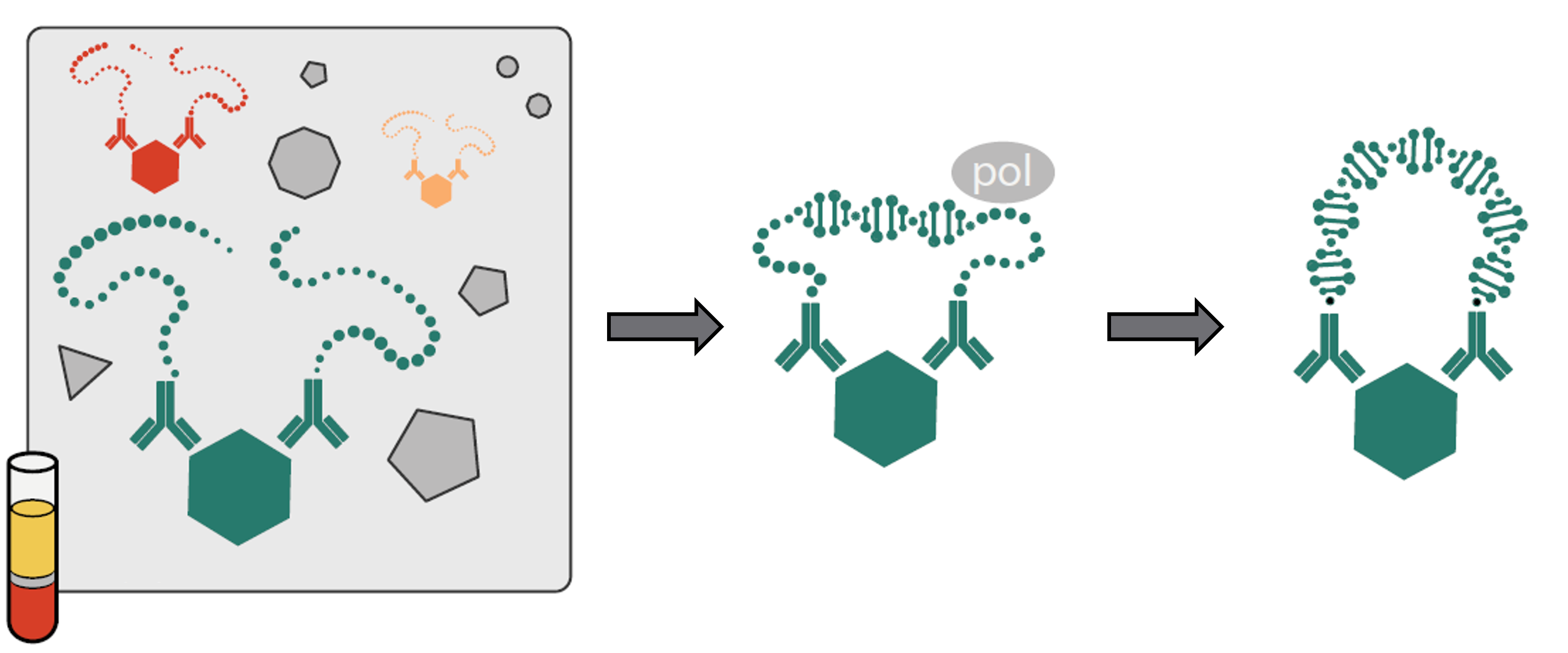
The Proximity Extension Assay (PEA) technology merges an antibody-based immunoassay with polymerase chain reaction (PCR). The basis of PEA is a dual-recognition immunoassay, in which two matched antibodies, labelled with unique DNA–tags, simultaneously bind to a target protein in solution. This brings the two antibodies into proximity and allows their DNA oligonucleotides to hybridize, which subsequently serve as template for a DNA polymerase–dependent extension step. The doublestranded DNA “barcode” thus generated uniquely represents the specific antigen and its abundance is quantitatively proportional to the initial concentration of target protein. Hybridization and extension are followed by PCR amplification, during which all extension products are amplified at identical rates and thus remain representative of the initial antigen concentration.
Matched antibody pairs (two monoclonal or a mix of polyclonal antibodies split into two fractions) are linked to different oligonucleotide tags, creating forward and reverse probes. Two different epitopes need to be recognized to achieve signal generation. This rendcers PEA technology superior to classical muliplex assays, as cross–reactivity is minimized.

How can you benefit from PEA technology?
In clinical research, proteomic analysis of complex human sample material like body fluids is a daunting task. The high dynamic range of protein concentrations [up to 12 orders of magnitude in human plasma; Geyer et al. (2017)] renders it challenging for conventional proteomic technologies to deliver a comprehensive picture of proteins at the whole range of concentrations and is frequently biased towards highly abundant proteins. In human plasma this problem is vividly exemplarixed by the fact that the 22 most abundant proteins comprise 99% of the whole protein content, which impairs investigation of other proteins of interest, in particular low abundant targets like cytokines or growth factors, which sharing the remaining 1% of the total protein concentration. Affinity proteomics approaches in general, and PEA in particular, are extremely effective to detect such low abundant proteins even in the context of highly complex samples with vast dynamic range of protein concentration as serum or plasma.
These challenges notwithstanding and due to their easy accessibility using minimally invasive methods rendering them monitorable in routine settings, proteomic analysis of body fluids remains interesting. This holds especially true for biomarker studies, as markers identified in these matrices bear the potential for immediate application in clinical settings for e.g. early disease detection, treatment responder stratification or to predict disease progression.
Development of suitable markers follows a multi-step pipeline in which high–throughput clinical discovery studies, analyzing a broad range of possible targets provide de novo biomarker profiling. Due to its superior performance in high dynamic range samples and thus body fluid, PEA technology frequently outperforms more conventional MS–based proteomics approaches. Additionally, the assay provides a high degree of multiplexing, as well as high specificity based on the requirement of target recognition by two complementary antibodies. The workflow is largely automated, ensuring precise liquid handling of minute volumes so that only small amounts of frequently precious clinical samples are consumed. Low abundant proteins can be detected in concentrations in the range of femtograms in parallel to measuring higher abundant proteins in the µg/mL range.
Advantages:
• High specificity: Dual-antibody recognition
• Wide dynamic range: fg/mL to µg/mL
• High throughput & minimal sample consumption: Automated workflows
• High multiplexing levels: ~5000 assays per sample in one run, 86(172) samples per run
Olink Explore
Olink Explore is a high–throughput biomarker discovery platform that combines PEA technology with a readout by Next Generation Sequencing (NGS). The flagship Olink Explore HT assay thus allows for the parallel analysis of 5416 unique proteins from minute amounts of liquid sample.
The platform is primarily designed for the analysis of serum and plasma samples, but other sample matrices are possible and have been validated by Olink. At the Core Facility Translational Proteomics we offer Olink Explore HT, as well as the smaller Explore 3072 and Reveal assay formats. The latter comprises 1000 target proteins at an attractive price point with an emphasis on immunologically relevant targets.
Lists of the relevant assay targets may be retrieved from Olink:
• Explore HT
• Explore 3072
• Reveal
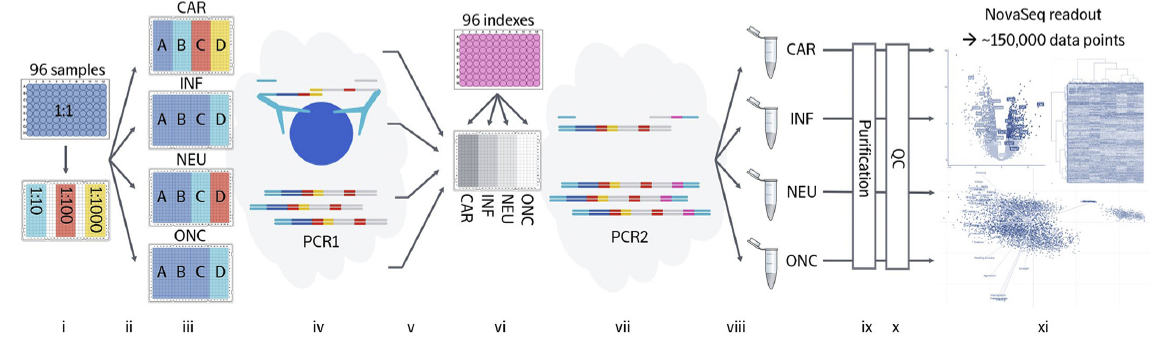
The Olink Explore Platform provides a semi–automatic workflow. Critical steps are performed using high–precision liquid–handling robots. This ensures consistent results and quality. The workflow comprises initial production of sample dilution series, dependent on sample matrix and panels to analyze. Samples are then incubated with the respective PEA probes, subsequent to which a first PCR is used for pre–amplification of the DNA–barcodes. In the next step, sample dilutions are pooled and combined with sample index primers that are appended to the amplicons in a second PCR. This results in DNA–barcodes unique to each assay and sample. Finally, the amplicons are pooled into libraries to be analyzed via NGS.
The platform encompasses targets with a broad dynamic concentration range and enables the analysis of both low and high abundance proteins (fg/mL – mg/mL).
To ensure the generation of high–quality data and data normalization, three internal and three external controls are included.
More information on the technology is available at the official Olink Website
Service
The Core Facility Translational Proteomics offers the Olink Explore technology to internal and external customers and collaborators. Apart from carrying out the measurements, we provide in–depth consultation regarding study design and execution. Our substantial expertise in statistical and bioinformatic data analysis is further available to accessibilize raw study output and aid with hypothesis generation.
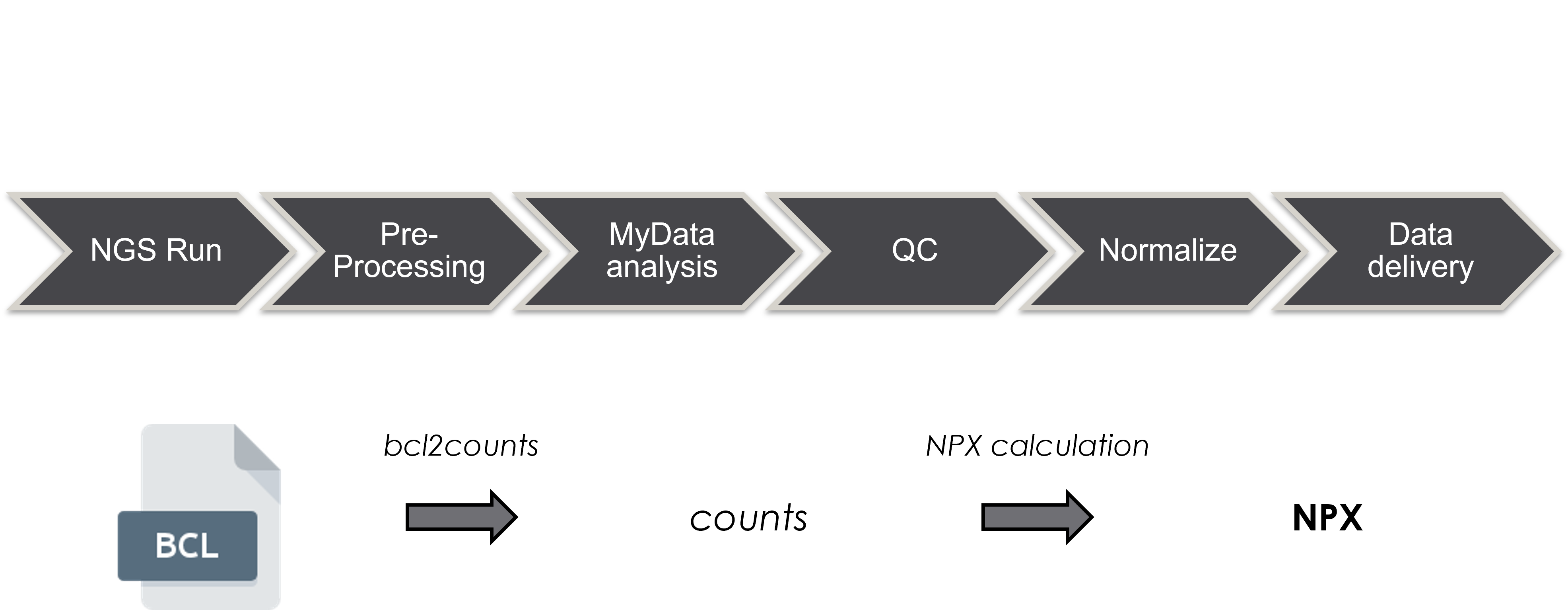
Data is made available as flat files containing Normalized Protein eXpression (NPX) values. Further, a Report of Analysis, containing information on intra– and inter–assay CVs, as well as the LOD for each assay, will be provided.
Should you be interested in harnessing Olink’s Explore affinity proteomics platform for your own research question, please do not hesitate to contact us for an initial discussion of your project and how we may provide the best service for your study.
Files Download
Sample Submission Form
Sample Submission Sheet 96 – well
Sample Submission Sheet 9x9 box
Sample Submission Sheet 10x10 box